Overview
Inside of each cell in your body, more than two meters of DNA is compressed into a space a few micrometers in diameter, an amazing amount of compaction. Our lab studies the packing of DNA in a variety of different contexts.
Our current research uses a combination of small-angle x-ray scattering (SAXS), inductively-coupled plasma atomic emission spectroscopy (ICP-AES), and other biophysical tools to study the electrostatic mechanism of DNA-DNA aggregation, the packing of DNA into viruses, and nuclesome self-attraction.
DNA Aggregation
A major part of our lab's research focuses on DNA which, in addition to being the blueprint for life, has one of the highest charges of any biomolecule. It is packed tightly in our cells (as mentioned above more than two meters of DNA is compressed into a space a few micrometers in diameter). Packing a molecule this long in such a small space is an interesting problem in its own right, but the highly-charged nature of DNA makes this an even more interesting problem. The large, negatively-charged DNA molecules should always strongly repel one another (like charges always repel one another). However, DNA does not always self-repel. Under certain solution conditions, namely solutions consisting of a small amount of +3 or higher ions, the DNA moves from a repulsive regime to a strongly self-attracting regime (See, for instance, Pelta et al., JBC, 1996). The DNA then aggregates (clumps together) and falls out of solution. Even more surprising is that adding even more of the exact same +3 or higher ions causes the DNA to move from the self-attracting regime back into the repulsive regime and redissolve. This is known as reentrant condensation or reentrant aggregation. Although the underlying physical forces are simple (the equations used to describe the system are similar to the ones we teach our sophomore physics students), how these forces combine to create this unexpected behavior is still a hotly debated topic.
Our research focuses on the positive and negative ions that are bound to the DNA in these aggregates and how these ions affect this aggregation. It builds upon previous work looking at the competition of ions of different valences (+1, +2, +3) in solution DNA (Andresen et al., PRL, 2004) and later work simultaneously measuring the binding of ions of various valences to DNA while monitoring the interaction between these DNA (Andresen et al., PRL, 2008}. We are using similar methods to look at the number and type of ions in the DNA aggregate. This comprehensive measurement of the ions has never been made and will give us the ability to distinguish between the various models that seek to describe this aggregation process.
Nucleosome Compaction
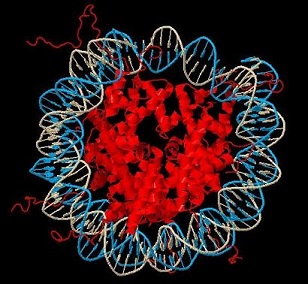
To acheive tight packing, DNA in our bodies is wrapped around the outside of a positively-charged, cylindrical complex of proteins, known individually as histones, to make the nucleosome core particle (NCP) (shown at left). The second step in this packing involves the tight packing of the NCPs against one another, a process that is still poorly understood.
What is known is that the NCPs can be made to attract each other by a change in solution conditions. A transition to a higher number of positively charged ions collapses free NCP monomers into large aggregates. The mechanism for this attraction, however, is still unclear. The problem is made more complex by the fact that the histones contain positively charged "tails" that extend outside of the negatively charged NCPs. Current theories suggest these tails are largely responsible for the electrostatic attraction between the NCPs. However, there has been little-to-no research on the (obviously key) role that the ions play in the interactions between these tails, the DNA, and the NCPs as a whole.
Our research looks at the role of these ions and the attractive forces generated between NCPs. Major questions include whether ions get released from the NCPs during this attractive phase or whether the ions are present in relatively constant concentrations throughout the process. What is the role of the positively-charged tails in the aggregation process? Does the NCP-NCP attraction mechanism have any similarities to the well-known phenomenon of DNA-DNA attraction? This research has enormous implications, as the transition of our DNA from loosely packed DNA to tightly packed DNA is a critical step in the transition of stem cells into normal cells. If we understand this process, we will be one step closer to being able to reverse this process and perhaps teach our cells to heal themselves! See information about how the "loosening" of DNA might be responsible for cell reprogramming in Science's Breakthrough of the Year. (Note: You or your institution need a subscription to Science to see this.)
Recent Publications
Quantitative Analysis of Single-Molecule Force Spectroscopy on Folded Chromatin Fibers
Nucleic Acids Research, 43(7) , 3578-3590 (2015). [html][pdf]
Solution Scattering and FRET Studies on Nucleosomes Reveal DNA Unwrapping Effects of H3 and H4 Tail Removal
PLoS One, 8(11) , e78587 (2013). [html][pdf]
Ion Competition in Condensed DNA Arrays in the Attractive Regime
Biophysical Journal, 105 , 984-992 (2013). [html][pdf]
Elucidating Inter-Nucleosome Interactions and the Roles of Histone Tails
Biophysical Journal , 105 , 194-199 (2013). [html][pdf]
*corresponding author