Primer Design Worksheet
The specificity of PCR
depends on the primers used to amplify a given segment of DNA. These primers
are on opposite sides, and strands, or the target fragment. Good primers are specific to a single
DNA sequence. For eDNA this
is particularly important as our DNA extraction contains many microbial genomes
from an environmental sample. In this lab, we want to only amplify a short DNA
sequence from the spotted salamander: Ambystoma
maculatum.
We will be designing primers
based on the A. maculatum cytochrome b gene.
You can view the record for
this accession here: http://www.ncbi.nlm.nih.gov/nucleotide/118202038
Š How long is this sequence? How many “bp?”
Š Note that this gene is found outside the nucleus. What
organelle has its own DNA?
Š When was the sequence published? What was the title of
the paper?
Š What is the string of letters following “translation=”
represent?
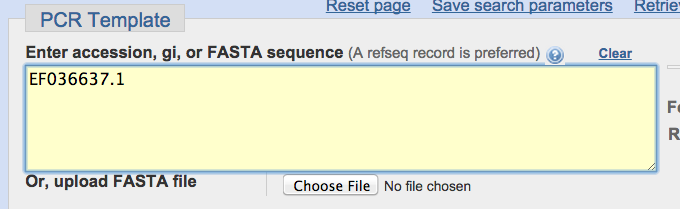
The Pubmed
accession number is: EF036637.1
Copy and paste this sequence
into the “Enter accession” box at the top of the primer design site:
http://www.ncbi.nlm.nih.gov/tools/primer-blast/
Under “Database” choose “nr.”
This is all published nucleotide data.

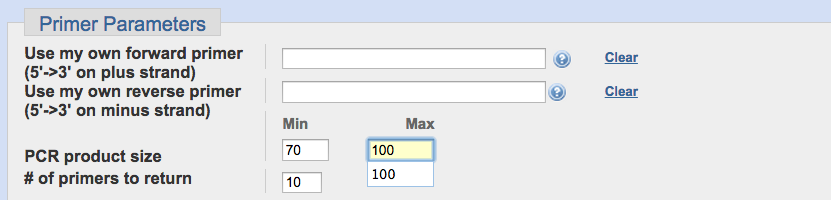
Under “Primer Parameters”
change the PCR product size “max” from “1000” to “100”
Under “Organism” we will add
organisms from the aquatic environment of out spotted salamanders. First click “Add
more organisms” then add: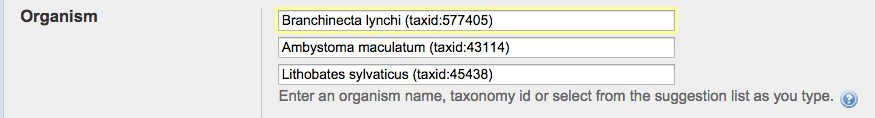
Ambystoma maculatum (taxid:43114)
Branchinecta lynchi (taxid:577405) (this is the “fairy
shrimp” that is often found in the same vernal pools)
Lithobates sylvaticus (taxid:45438)

Fairy shrimp
Under “Primer specific
stringency” change the “total mismatches from 2 to 3.
We will leave everything else
as a default.
Click: ![]()
You will be prompted with the
following.
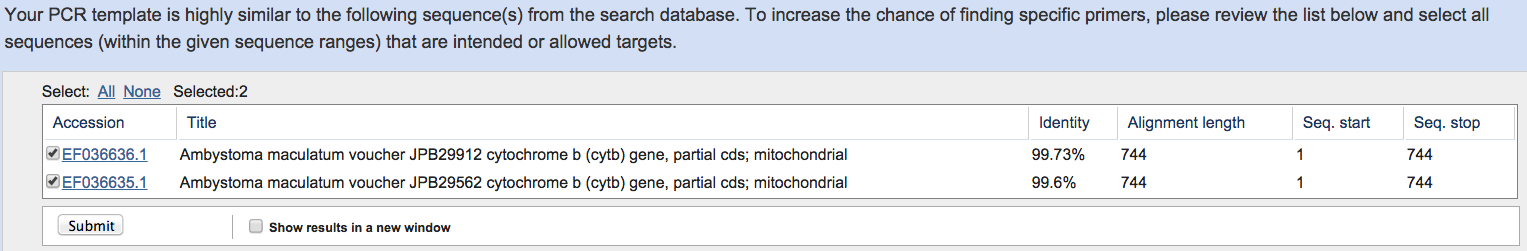
Select both of these Cyt B submissions and hit “Submit”
Your results should look like
this:
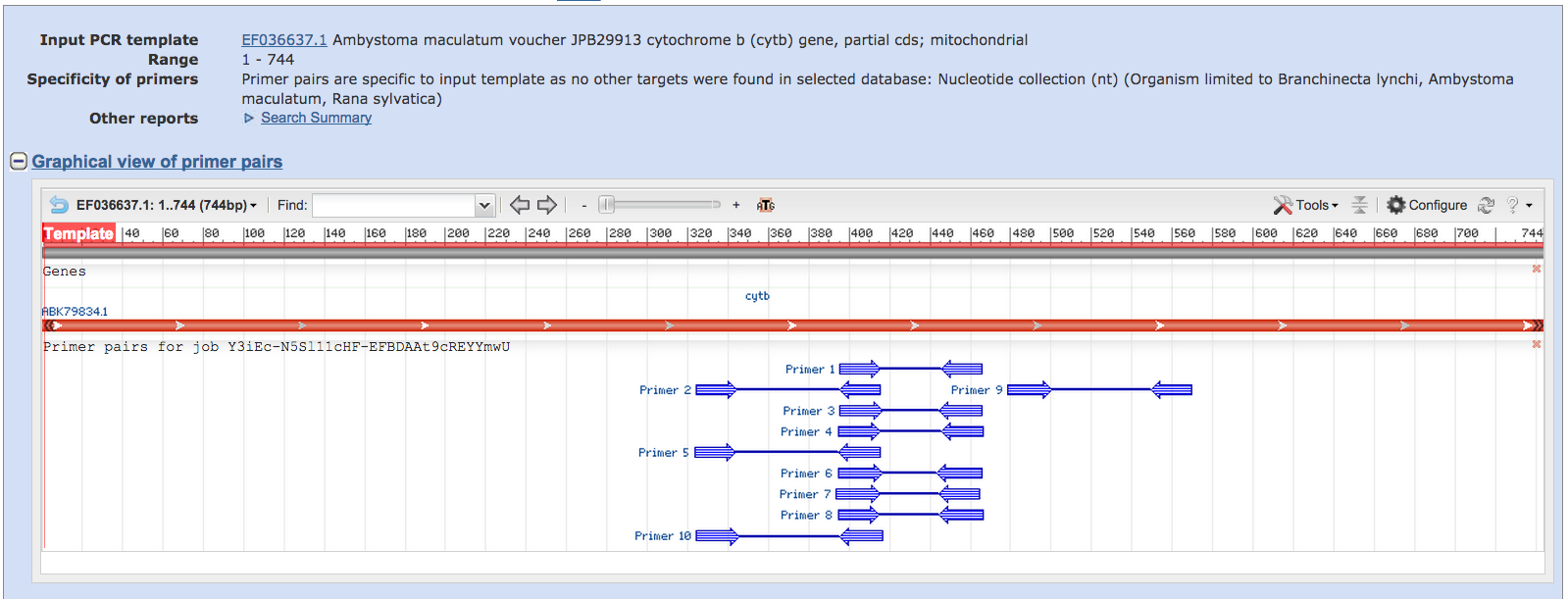
The top “Template” bar is a
graphical representation of your submitted sequence. The program has returned
10 primer pairs.
Scroll through the Detailed
Report.” Note that none of the entries bind to published
fairy shrimp or wood frog sequences.
Each report contains the
following:
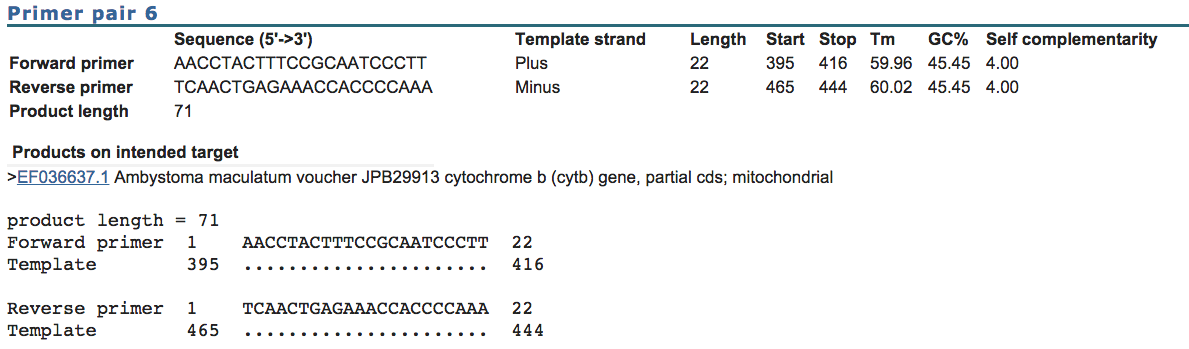
Note the forward and reverse
primer sequences bind to the two different strands (plus and minus) and in this
example each has a length of 22bp.
The distance between the
beginning of the forward primer and beginning of the reverse primer is the
product length (465-395 = 71bp).
The “Tm” s
the predicted annealing temperature of this primer to the target DNA sequence.
The GC% is the percentage of guinines and cytosines. We
typically like something around 50%.
The “self complimentarity”
score is a measure of how likely the primer is to fold on itself. We like to
see values below 6.00.